Dionysus
Performance, calibration, and generalizability of probabilistic models for small chemical datasets.
Published 02/05/2023 in Digital Discovery (Royal Society of Chemistry)
This was work done with Riley J. Hickman, Aniket Zinzuwadia, Afshan Mohajeri, Benjamin Sanchez-Lengeling and Alán Aspuru-Guzik.
Link to paper is here, and link to the Github repo is here.
In this work, we focus on the performance, calibration, and generalizability of probabilistic models when they are trained on low-data experimental chemical datasets. Datasets range from 100-2000 data points, similar to the data availability of early chemical discovery campaigns. A wide variety of models were testing, including tree-based (NGBoost), kernel based (Gaussian process), and deep probabilistic models (BNN, MLP-GP, and GNN-GP). Four commonly used featurization methods are used: circular fingerprints, Mordred physicochemical descriptors, GNN embeddings, and graph representation. We study the performance and calibration–the reliability of uncertainty estimates provided by the probabilistic model–for the various tasks, featurizations, and models.
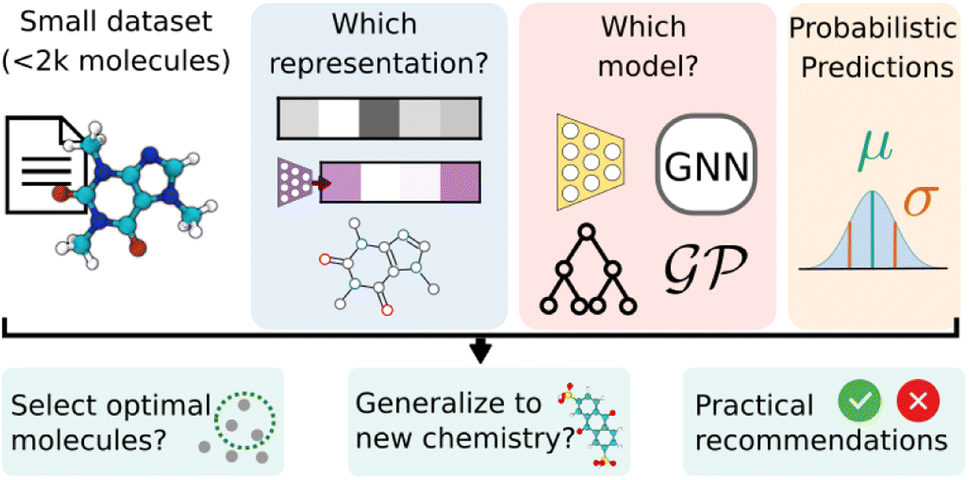
Additionally, the models were tested as surrogates for a simulated Bayesian optimization campaign, with the goal of identifying the optimal compound in the dataset. We introduce “cluster splits” for testing the generalizability of the models, simulating out-of-distribution datasets through the ablation of portions of the chemical space, as defined by the molecular structure, and target property. Finally, we make some practical recommendations for applying regression and classification machine learning models to small chemical datasets.