GAUCHE
Open-source package for GAUssian processes for CHEmistry.
Published 19/12/2023 in NeurIPS 2023
This was work done with Ryan-Rhys Griffiths, Leo Klarner, Henry Moss, Aditya Ravuri, Sang T. Truong, Yuanqi Du, Samuel Don Stanton, Bojana Ranković, Arian Rokkum Jamasb, Aryan Deshwal, Julius Schwartz, Austin Tripp, Gregory Kell, Simon Frieder, Anthony Bourached, Alex James Chan, Jacob Moss, Chengzhi Guo, Johannes P. Dürholt, Saudamini Chaurasia, Ji Won Park, Felix Strieth-Kalthoff, Alpha Lee, Bingqing Cheng, Philippe Schwaller, Jian Tang and Alán Aspuru-Guzik.
Link to paper is here, and link to the Github repo is here.
Predictive tasks are important for materials and drug discovery research. While more complex models (ie. deep learning models) have demonstrated state-of-the-art performance on regression and classification tasks, kernel methods such as Gaussian processes are still commonly used, due to the low amounts of data available for training when working with experimental datasets. This is usually due to the costly and resource-intensive nature of performing the experiments.
GAUssian processes for CHEmistry (GAUCHE) is a library for working with molecules, chemical reactions, and proteins. In this library, we provide a series of representations for the various inputs, along with custom kernels use for Gaussian processes, which include: bit-wise fingerprint kernels, string kernels, and graph kernels. The package is built on top of PyTorch, GPyTorch and BOTorch.
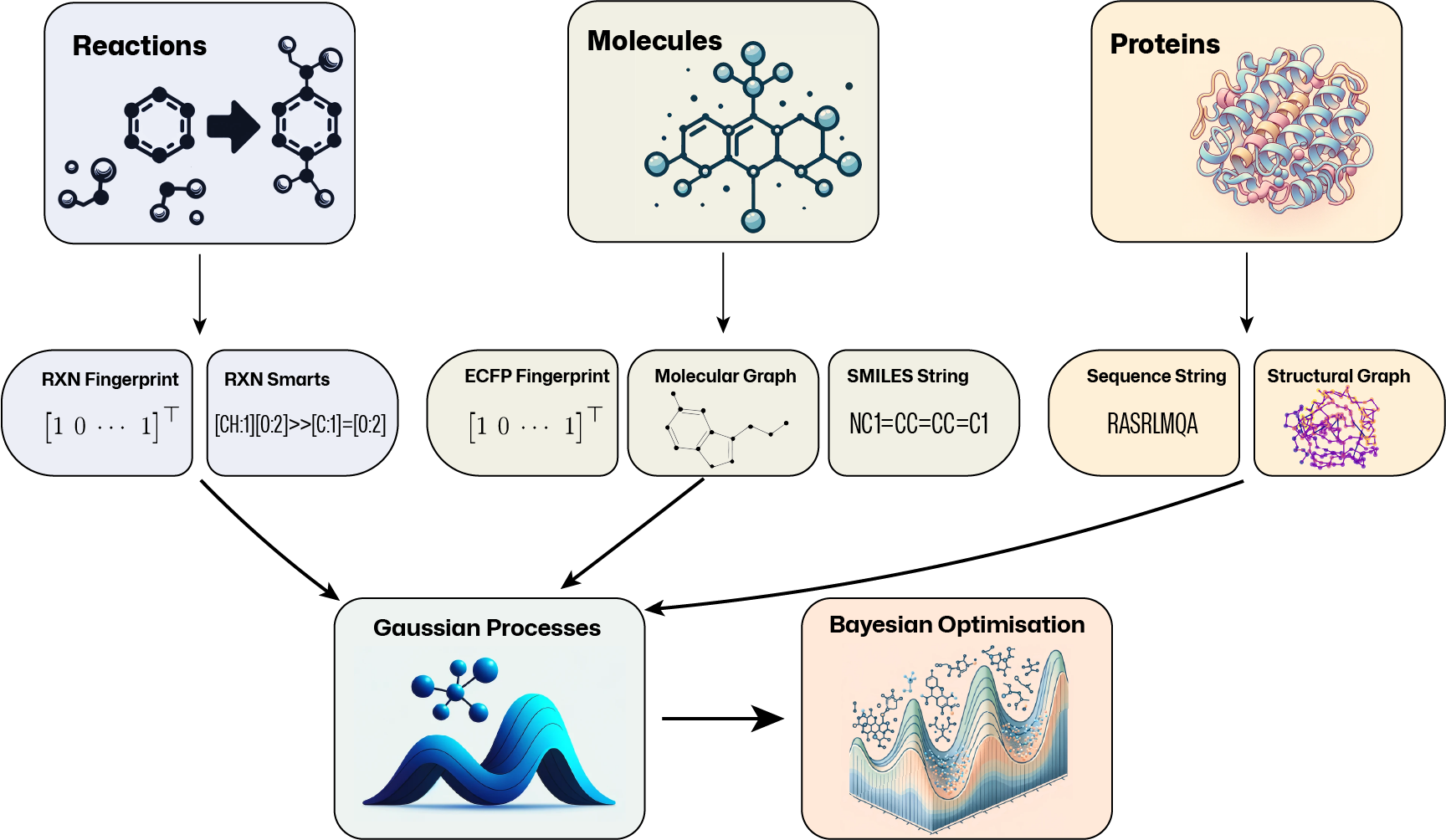
The performance and calibration of the kernels/representations, along with results with Bayesian neural networks, are compared for the different tasks and datasets. We also provide analysis on the Bayesian optimization performance of the kernels/representations.